56
PNL Volume 15 1983
RESEARCH REPORTS
PEA 6-PHOSPHOGLUCONATE DEHYDROGENASE ISOZYMES
Weeden, N. F. NYS Agricultural Experiment Station, Geneva, NY, USA
Two isozymes of 6-phosphogluconate dehydrogenase (6PGD) have been
identified in spinach (1), radish (2), castor bean (4) and Senecio syl-
vaticus (5). In each of these species one of the isozymes is found in
the cytosol while the second is localized in the plastid (chloroplast,
leucoplast, etc.). In this report I present evidence that two isozymes
of 6PGD are also present in pea leaves, that one of these is in the
cytosol and the other in the chloroplast, and that the two isozymes are
specified by distinct genes.
Starch gel electrophoresis was performed as described in the accom-
panying article (8) using the pH 6.1 buffer system. Chloroplast and
cytosolic fractions were obtained as described previously (6) . The as-
say for 6PGD was modified from Shaw and Prasad (3). Two zones of 6PGD
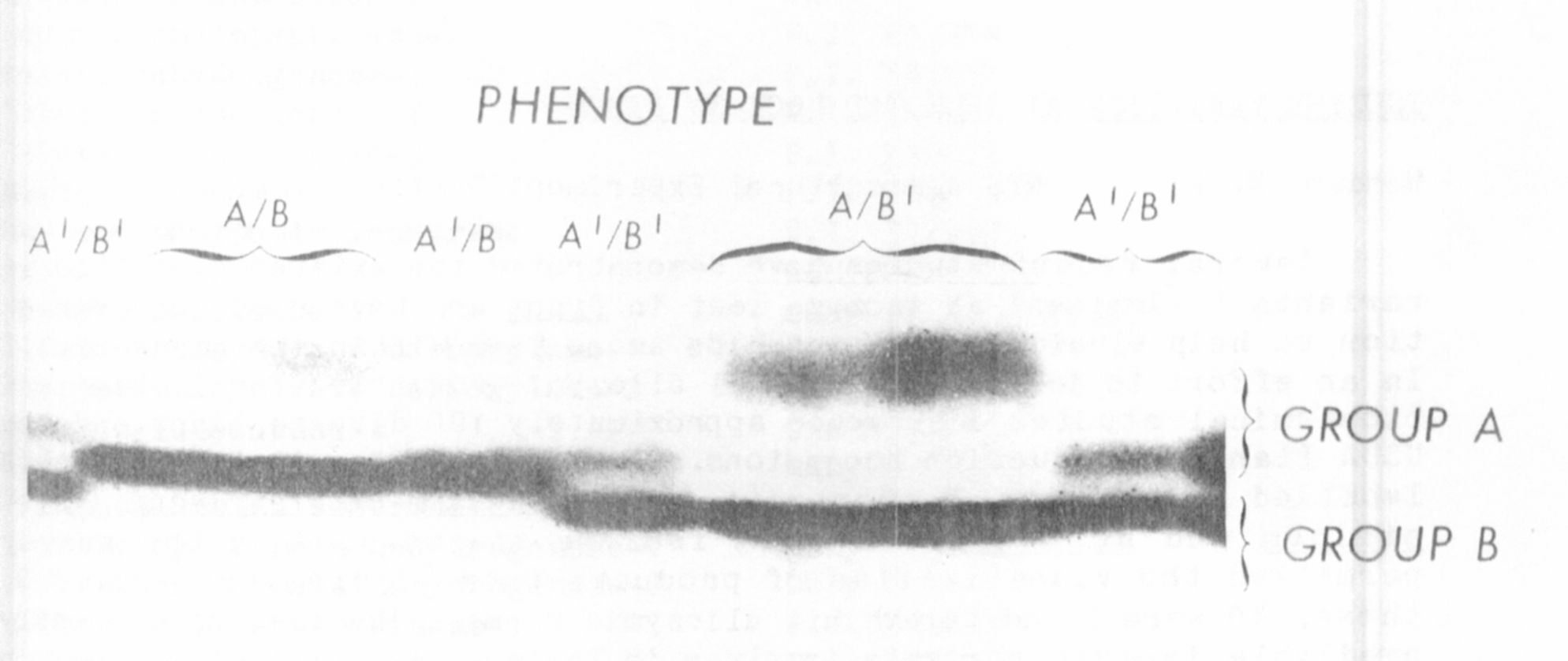
activity were observed after electrophoresis of extract from a number of
inbred lines (Fig. 1). In every line examined only one band of activity
within Group A and one within Group B were observed (see Fig. 1). The
more anodal set (Group A) was present in whole leaf and chloroplast ex-
tracts but absent from soaked pollen extracts, indicating a plastid
localization. The slower migrating bands (Group B) were observed in
whole leaf and soaked pollen but not in the chloroplast pellet and are
believed to be contained in the cytosol. Mitochondrial, peroxisomal and
vacuolar compartments were not investigated.
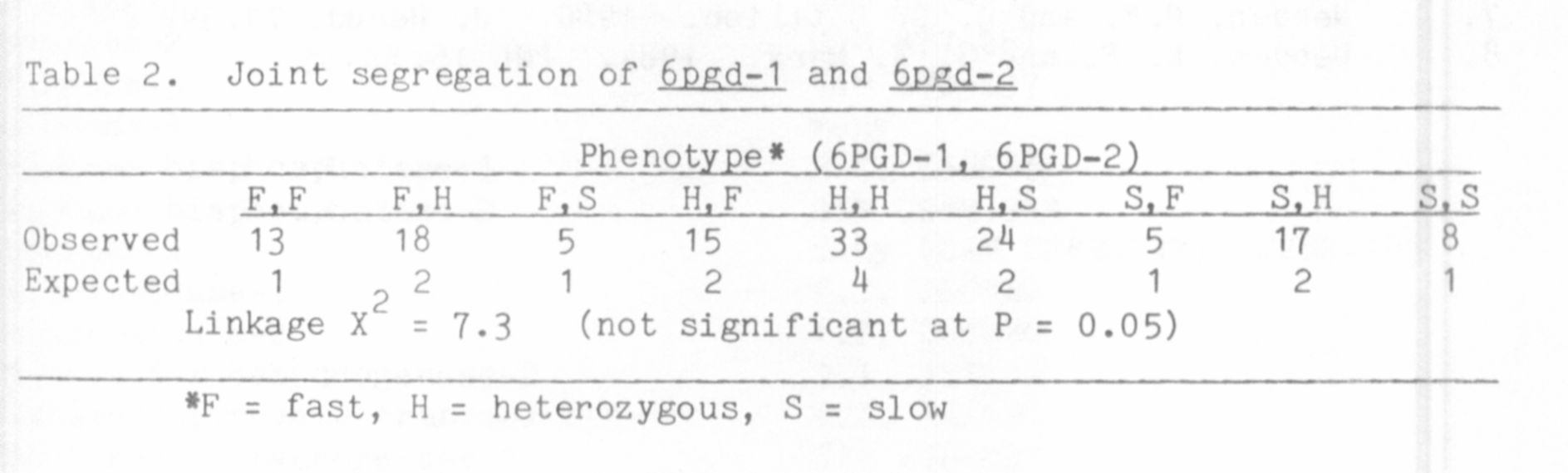
The variability present between lines permitted genetic analysis of
the isozymes by classical inheritance studies. An appropriate F2
progeny was tested for segregation of forms A and A' and forms B and B'.
Within each set three phenotypes were observed in the F2; two were
parental and the third consisted of a broad region of activity covering
both parental zones. This third phenotype was that seen in all F1
plants and is considered to be that of a heterozygous individual because
allelic forms of isozymes are generaly codominant. For both sets the
ratio of the three phenotypic classes in the F2 was very close to the
1:2:1 ratio expected for two alleles segregating at one locus (Table 1).