
Inverse
PCR to identify DNA sequence upstream
of
the pea HMG I/Y open reading frame
Druffel,
K.,
Hartney,
S. and Hadwiger,
In the pursuit of the defense responses in pea plants, or in any induced response in eukaryotic organisms, the sequence of the open reading frame of the induced gene is attainable from a cDNA clone. Once the open reading frame transcription start site of the cDNA clone has been obtained, the promoter region is often of interest and is accessible by inverse PCR. Although inverse PCR is a standard technique, all PCR strategies do not necessarily result in success. The promoter of the pea gene HMG I/Y is of interest because of the possibility that its coded protein is influencing its own transcription. The HMG I/Y protein contains four AT-hooks (2, 5) that have affinity for AT-rich regions of DNA (4). The extensive research relative to this property has led to the recognition that HMG-I/Y is an architectural transcription factor (7). It has recently been determined that both its RNA transcript and its protein product are depleted as the pea defense response is initiated (4).
The pea HMG I/Y protein can efficiently bind to AT-rich segments of promoters from pea PR genes, genes that are activated as the pea tissue resists fungal pathogens (1, 4). Therefore it was of interest to determine if such AT-rich regions were also present in the HMG I/Y�s 5' region. This manuscript describes in detail the strategy used to sequence this promoter.
Methods and Results
DNA extraction
One immature pea pod (3 cm length) from Pisum sativum was crushed between two sheets of weighing paper using pliers. The pod was then transferred to a 1.5 ml microfuge tube with 1 ml of extraction buffer (6) (100 mM Tris, pH 8.0, 50 mM EDTA, 500 mM NaCl, 10 mM 2-mercaptoethanol). 140 �l of 10% SDS was added. The tube was inverted to mix and incubated at 65 C for 10 minutes. 250 �l of 8M KOAc was added to the tube, inverted to mix, and placed on ice for 5 minutes. The tube was then centrifuged at 13,000 rpm for 8 minutes and 600 �l of the supernatant was transferred to a new tube. 300 �l of isopropanol was added and the contents were mixed and held at 4 C for 10 minutes. Following 10 minutes of centrifugation at 13,000 rpm the supernatant was discarded. 750 �l of 75% EtOH was added. The tube was gently mixed then centrifuged for 3 minutes. All supernatant was removed from the tube and the pellet resuspended in 50ul sterile ddH20.
Inverse PCR
Pea genomic DNA was cleaved with the TaqI
4 bp cutter (BRL products/Life Technology,
Ligated TaqI/cut genomic DNA 1.2 ul
10X PCR buffer (BRL) 2.2 �l
2.5 mM dNTPs 1.2 �l
50 mM MgCl2 0.8 �l
20 �m LeeInv367c (5� GAA CAA CCG AAT GGC CTT CT 3�) 0.6 �l
20 �m LeeInv580F (5� CCA AAG GCT TCT GGA AGT GG 3�) 0.6 �l
double distilled H2O 13.6 �l
Taq Polymerase 0.2 �l
The PCR temperature recycling program was: 94 C for 30 sec, 65 C for 20 sec, 72 C 1.5 min for 50 cycles. The product was then diluted 1:10 in preparation for secondary PCR.
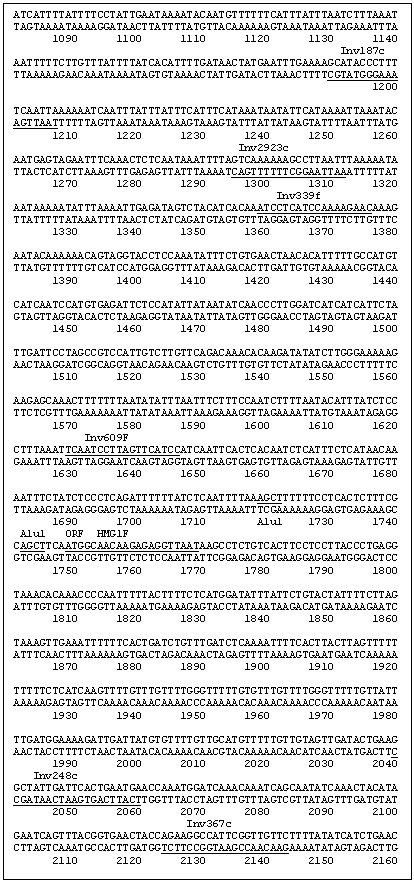
A secondary PCR utilized the primer combination INV615F (5' GCC GAA GAA GAT TGC TAG GAC 3')/INV248c (5' TCA TTC AGT GAA TCA ATA GCC 3') as indicated in Fig. 1 with an annealing temperature of 61 C. This yielded an ~780 bp segment, which was cloned into a Topo PCR2.1 vector (Promega) and subsequently sequenced. The two new sequences generated from the clone were recognized as being within the region 5' of the HMG I/Y start site. The length of this sequence (780 bp) was insufficient for the complete analysis of a HMG I/Y 5' region but contained an AluI site. Thus the pea DNA was cut with AluI and the DNA was again circularized by ligation to obtain additional upstream sequence. Two new sets of primers were designed. Inv339F (5' ATC CTC ATC CAA AAG AAG 3�)/ Inv292 (5' AAT TAA GGC TTT TTT GAC 3') was used for the primary PCR reaction. PCR was run under the following conditions; 94 C for 30 sec, 51 C for 20 sec, 72 C for 1 min. Secondary PCR using Inv187c (5' TAA TTG AAA AGG GTA TGC 3')/Inc609f (5' TCA ATC CTT AGT TCA TCC 3') was then performed (94 C for 30 sec, 51 C for 20 sec, 72 C for 1 min for 50 cycles). The subsequent PCR product was cloned and sequenced as before and increased the total sequence upstream of the HMG-I/Y gene to 1748bp (Fig. 1).
To verify the sequence 1748bp upstream of the HMG-I/Y gene, a primer set 13f (5� ACA GAT TAT GAA CAA AGT TTA ACG 3�)/754 (5� TCA CTT GTG TCA ACT GAG GC 3�) was used to amplify an approximate 2500 bp segment off of uncut genomic DNA. PCR conditions were 94 C 30sec, 61 C 20sec, 72 C 2:4 min for 50 cycles. The PCR product was cloned and completely sequenced to confirm that the inverse sequence reactions were assembled correctly.
Discussion
The availability of this sequence enables the identification of potential promoter elements 5' of the HMG-I/Y opening reading frame and assists the development of nested series of promoter/reporter elements to evaluate those sequences vital for activation (1). Once these regulatory elements have been found it is possible to identify and characterize the sequences involved in
HMG-I/Y transcription. Stretches of alternating A and T sequences are known to bind the AT hooks of the HMG-I/Y protein (7). There are fifteen 4 bp stretches, three 5 bp stretches, four 6 bp stretches and one 7 bp stretch of alternating A and T found in the region 5' of the pea HMG-I/Y gene open reading frame. The availability of pure HMG-I/Y protein and the 5' sequence information enables gel mobility assays. These assays can determine if any of the regions 5' of HMG-I/Y associate with this architectural transcription factor that is its own coded gene product. Many other predicted transcription factor binding sites can be derived from the 5' sequence. For example, the MYB attachment site, AACCG, is found twice in the pea 5' region and once in the Arabidopsis promoter region (3).
Acknowledgment:
The GenBank Accession number for the pea HMG-I/Y sequence reported in this
manuscript is AY864056. We thank the Washington Sea Grant program for support.
Ag.
1.
Choi, J.J., Klosterman, S.J. and Hadwiger,
2. Gupta, R., Webster, C.I. and Gray, J.C. 1997. Plant Mol. Biol. 35: 987-992.
3. Gupta, R., Webster, C.I. and Gray, J.C. 1998. Plant Mol. Biol. 36: 897-907.
4. Klosterman, S. J., Choi, J. J.,Hadwiger, L. A. 2003. Mol. Plant Path. 4: 249-258.
5. Klosterman, S.J. and Hadwiger, L.A. 2002. Plant Sci. 162: 855-866.
6. Presting, G.G., Smith, O.P. and Brown, C.R. 1995. Phytopath. 85:436-442.
7. Reeves, R. 2001. Gene 277: 63-81.



Fig. 1: DNA sequence of the 5' region and the open reading frame of the pea gene HMG-I/Y. The restriction enzyme sequences and the primers utilized in developing the 5' sequence are underlined and labeled.
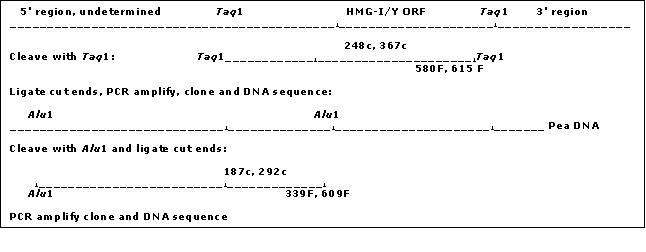
Fig. 2 Cartoon of the sequence of restriction digestion, primer development and DNA sequencing analyses of the pea HMG-I/Y 5' region (See Methods).