genotypic sequence polymorphism was defined (6). PCR primers were designed to distinguish gene polymorphisms between pea lines having high or low TIA, and various PCR patterns were described for a number of lines (6). These patterns were scored first on 95 lines from a collection of pea genetic resources, where a correlation between a 646 bp PCR product and low TIA values was observed (6). We now examine the utility of these markers for breeding programs and report the results of testing this set of PCR primers on a population of 168 RILs from a cross between two pea lines having high or low TIA.
When the population of RILs derived from parents having high or low TIA was screened for PCR product size and TIA, in no case did the 646 bp band result from a line with a high TIA (Figure 1). PCR markers based on the Tri locus are thus linked to TIA in pea seeds, providing strong markers directly located to TI genes and based on a simple PCR reaction. This type of marker promises to be of enormous value to breeders, especially if the DNA extracted from populations can be screened for additional markers simultaneously.
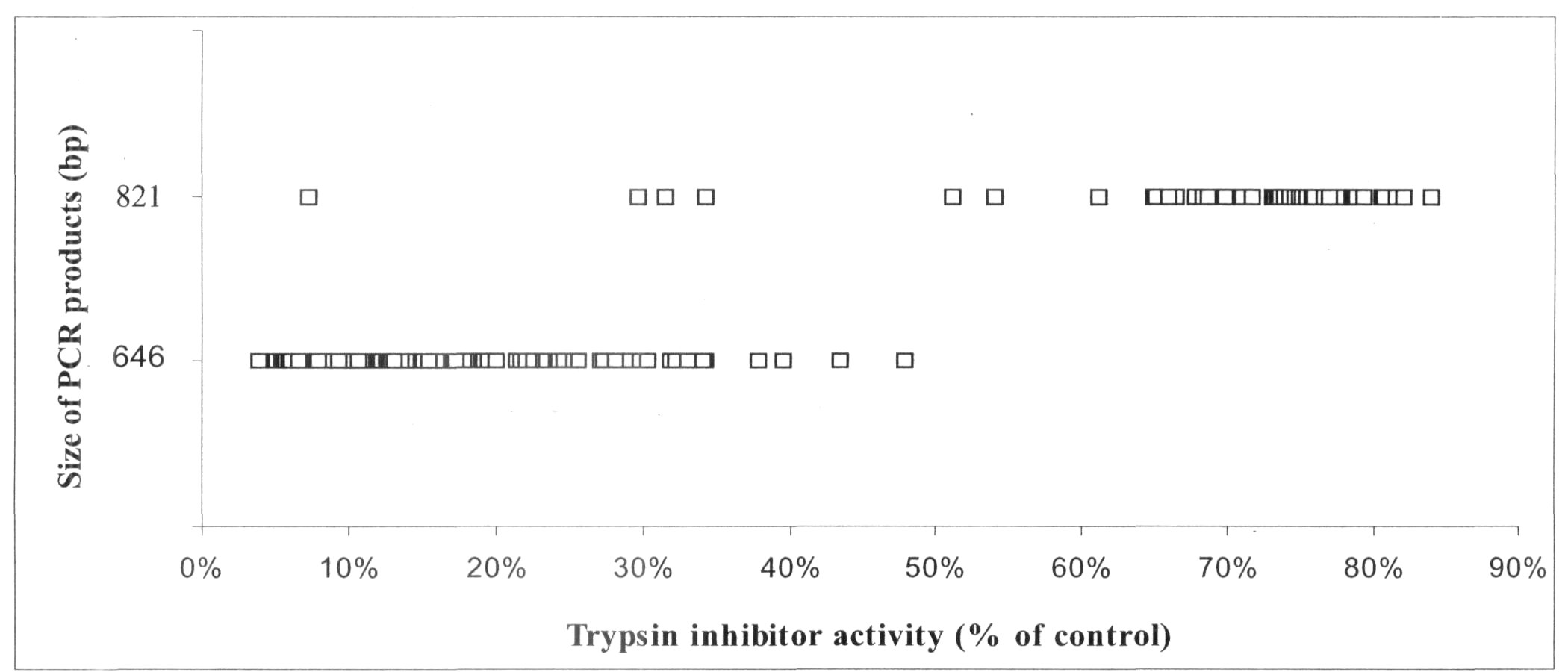
Fig. 1. Summary of PCR results for a population of 168 recombinant inbred lines from the cross Terese (low TIA) X Champagne (high TIA). Scores for two PCR bands of 646 and 821 bp are shown (ordinate). The TIA of each line is indicated on the abscissa; the higher the percentage, the higher the corresponding TIA.
The strategy of marker-assisted breeding promises to be valuable because of potential saving in the cost of phenotyping/genotyping. In some cases, it can also reduce errors due to environmental effects on gene expression. We illustrate here how knowledge of genes can be used to provide simplified tools to breeders that can be used to facilitate selection of variants for legume seed composition. The present rapid development of finer genetic maps, with new types of markers, as well as the development of robotics for genomics, should make molecular approaches for breeding even more efficient in the future. Selection may then be based on markers that are very close to the character and, in addition, numerous seed compounds may be screened simultaneously.
Acknowledgements: UNIP (Paris, France) and DEFRA (UK) are acknowledged for financial support for this work.
1. Domoney, C, Welham, T., Ellis, N. and Hellens, R. 1994. Theor. Appl. Genet. 89: 387-391.
2. Domoney, C, Welham, T., Sidebottom, C. and Firmin, J. 1995. FEBS Lett. 360: 15-20.