Mapping
of the breakpoints of the Twt-translocation
Gorel, F.L, and Berdnikov, V.A.
Inst. of Cytology and Genetics, Russian Academy of Sciences,
Novosibirsk, Russia
 In 1992 we described a dominant mutation Twt
(Twisted tendrils) found in the M2
population after treatment of seeds of the line Sprint2 with EMS (1).
Surprisingly, this mutation appeared to be connected with one of the
breakpoints of a new reciprocal translocation found in the same plant and which
most probably arose from the same mutation event. In subsequent experiments we
have not observed the translocation without
Twt. Thus, the Twt mutation is a marker of the new translocation, which we
called Twt-translocation. We have
shown that the locus Twt resides on
the long arm of chromosome II (for simplicity, here we use the numerals of
linkage groups to designate chromosomes), about 8 cM from the locus A
towards the telomere (1, 2). Later we found pseudo-linkage (18% crossing-over) between Twt
and the locus D (4), a marker on chromosome I not far from the telomere (5).
Thus, the Twt-translocation involves chromosomes I and II.
In 1992 we described a dominant mutation Twt
(Twisted tendrils) found in the M2
population after treatment of seeds of the line Sprint2 with EMS (1).
Surprisingly, this mutation appeared to be connected with one of the
breakpoints of a new reciprocal translocation found in the same plant and which
most probably arose from the same mutation event. In subsequent experiments we
have not observed the translocation without
Twt. Thus, the Twt mutation is a marker of the new translocation, which we
called Twt-translocation. We have
shown that the locus Twt resides on
the long arm of chromosome II (for simplicity, here we use the numerals of
linkage groups to designate chromosomes), about 8 cM from the locus A
towards the telomere (1, 2). Later we found pseudo-linkage (18% crossing-over) between Twt
and the locus D (4), a marker on chromosome I not far from the telomere (5).
Thus, the Twt-translocation involves chromosomes I and II.
For a more precise location of the breakpoint on chromosome I we crossed
our line Twt-T (being a structural homozygote for
Twt-translocation originating from cross VIR3953 x Sprint2 Twt/twt
described in ref. 1) with line WL6115 (a,
twt, brac). The locus Brac shows
linkage to D (27.5 % of crossing-over,
ref. 3). It should be noted that
among plants with Twt phenotype the
homozygotes Twt/Twt can be easily
distinguished from heterozygotes Twt/+
by pollen fertility, heterozygotes exhibiting 50% sterile pollen.
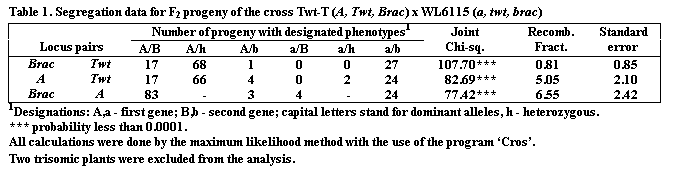
The data presented in Table 1 show that all the three segregating loci, A,
Twt and Brac, are tightly linked.
The strongest linkage (0.8%) is found between the loci Twt
and Brac.
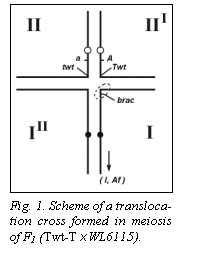
Fig. 1 shows the presumed position of the recessive allele
brac on chromosome I with a dashed oval.
 A more precise location of Brac was
achieved by trisomic analysis. Earlier (1) we have shown that in the progeny of
structural heterozygotes for the Twt-translocation
there occur tertiary trisomics that possess a standard diploid karyotytpe plus a
small interchange chromosome III. In the F2 population
analyzed, we found two such trisomics (their karyoptype was supported by
analysis of PMC). The plants
displayed the phenotype A Twt brac.
Thus, the recessive allele brac was
not covered by the extra chromosome III.
Instead, it is situated on chromosome I more proximally (lower on the
diagram) than the translocation breakpoint.
A more precise location of Brac was
achieved by trisomic analysis. Earlier (1) we have shown that in the progeny of
structural heterozygotes for the Twt-translocation
there occur tertiary trisomics that possess a standard diploid karyotytpe plus a
small interchange chromosome III. In the F2 population
analyzed, we found two such trisomics (their karyoptype was supported by
analysis of PMC). The plants
displayed the phenotype A Twt brac.
Thus, the recessive allele brac was
not covered by the extra chromosome III.
Instead, it is situated on chromosome I more proximally (lower on the
diagram) than the translocation breakpoint.
Acknowledgement: This
work was supported by the Russian State Program 'Russian Fund for Fundamental
Research', grant No 99-04-49970.
1. Gorel,
F. L., Temnykh, S. V., Lebedeva, I. P., and Berdnikov, V. A. 1992. Pisum
Genetics 24: 48-49.
2. Gorel,
F. L., Kosterin, O. E., and Berdnikov, V. A. 2000. Pisum Genetics 32: 57-58.
3. Rozov,
S. M., Gorel, F. L., Berdnikov, V. A. 1997. Pisum Genetics 29: 26.
4. Temnykh,
S. V., Gorel, F. L., Berdnikov, V. A., and Weeden, N. F. 1995. Pisum Genetics
27: 23-24.
5. Murphy,
R.L., Weeden, N.F., and Przyborowski, J.A. 2000. Pisum Genetics 32: 39-41.
 In 1992 we described a dominant mutation Twt
(Twisted tendrils) found in the M2
population after treatment of seeds of the line Sprint2 with EMS (1).
Surprisingly, this mutation appeared to be connected with one of the
breakpoints of a new reciprocal translocation found in the same plant and which
most probably arose from the same mutation event. In subsequent experiments we
have not observed the translocation without
Twt. Thus, the Twt mutation is a marker of the new translocation, which we
called Twt-translocation. We have
shown that the locus Twt resides on
the long arm of chromosome II (for simplicity, here we use the numerals of
linkage groups to designate chromosomes), about 8 cM from the locus A
towards the telomere (1, 2). Later we found pseudo-linkage (18% crossing-over) between Twt
and the locus D (4), a marker on chromosome I not far from the telomere (5).
Thus, the Twt-translocation involves chromosomes I and II.
In 1992 we described a dominant mutation Twt
(Twisted tendrils) found in the M2
population after treatment of seeds of the line Sprint2 with EMS (1).
Surprisingly, this mutation appeared to be connected with one of the
breakpoints of a new reciprocal translocation found in the same plant and which
most probably arose from the same mutation event. In subsequent experiments we
have not observed the translocation without
Twt. Thus, the Twt mutation is a marker of the new translocation, which we
called Twt-translocation. We have
shown that the locus Twt resides on
the long arm of chromosome II (for simplicity, here we use the numerals of
linkage groups to designate chromosomes), about 8 cM from the locus A
towards the telomere (1, 2). Later we found pseudo-linkage (18% crossing-over) between Twt
and the locus D (4), a marker on chromosome I not far from the telomere (5).
Thus, the Twt-translocation involves chromosomes I and II.
