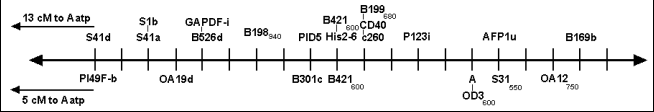
The position of locus A on the consensus map
Przyborowski, J. and Weeden, N.F. Department of Plant Sciences and Plant Pathology
Montana State University, Bozeman, MT 59717 USA
One of the most familiar and useful morphological markers in pea is the white-flowered mutation at the A locus. This gene has been involved in countless mapping experiments since Mendel first brought it to prominence as one of the seven traits he examined in his pioneering genetic studies. In the classical linkage map for pea published by Blixt in 1972 (2) the locus appeared as the terminal marker at the upper end of chromosome 1. The position of A has since been revised several times, and it is now known to be neither terminal nor on linkage group I. Instead, the most recent linkage map of pea places A and its closely linked markers on linkage group II (4). Unfortunately, gene a was not segregating in the JI1794 x Slow RIL population used to generate the backbone of the consensus map. Hence the precise position of this important marker could not be specified.
In order to address this problem we examined the location of A in an RIL population derived from the cross MN313 x JI1794. MN313 is a line homozygous a, and by crossing with JI1794 we can use many of the markers present on the consensus map to precisely localize the position of the A locus on the consensus map. Fig. 1 shows a comparison of the region around A for the maps generated for each of the RIL populations. The RAPD B421600 segregated and mapped in each set of RILs, as did Aatp. The calculated distance between B421e and Aatp is greater in the JI1794 x Slow RIL population (22 cM) than in the MN313 x JI1794 RIL population (14 cM). However, this difference is not significant, reflecting the relatively low precision in recombination frequencies when working with RIL populations of only about 50 lines. What is more critical is that this region is relatively well saturated on both maps and the order of the markers appears to be conserved. In the JI1794 x Slow RILs marker B421600 cosegregated with His2-6, a well-studied component of the genes encoding the histones of pea (3). The position of A on the map generated for the MN313 x JI1794 RILs is 5 cM from B421600 on the opposite side of the marker from Aatp. This position agrees well with other estimates of the position of A relative to His2-6 (3). It appears that the position of A on the consensus map is very close to the AFLP marker afp1u, mapped by Gail Timmerman-VaughanÆs group in Christchurch, New Zealand. Such comparative mapping using JI1794 as one parent of a cross should permit the placing of many other loci on the consensus map that are not segregating in the original JI1794 x Slow RILs.

Fig. 1. Sequence of markers in the region surrounding the A locus on the consensus map (above horizontal line) and on the map developed for the MN313 x JI1794 RIL population (below horizontal line). Cross hatch markings are spaced at 1 cM intervals. The position of the RAPD B421600, mapped in both popultaions, is approximately in the center of the figure. The position of A is 5 cM to the right of B421600 on the lower map. The locus Aatp is off the map on the left hand side of the figure for each map. The distance between B421600 and Aatp is different for each map, indicating the relative scales for the two maps differ slightly.
1. Berdnikov, V.A. and GorelÆ, F.L. 2000. Pisum Genetics 32:6-8.
2. Blixt, S. 1972. Agri. Hort. Genet. 30: 1-293.
3. Kosterin, O.E. 1992. Pisum Genetics 24: 56-59.
4. Weeden, N.F., Ellis, T.H.N., Timmerman-Vaughan, G.M., Swiecicki, W.K., Rozov, S.M. and Berdnikov, V.A. 1998. Pisum Genetics 30: 1-4.