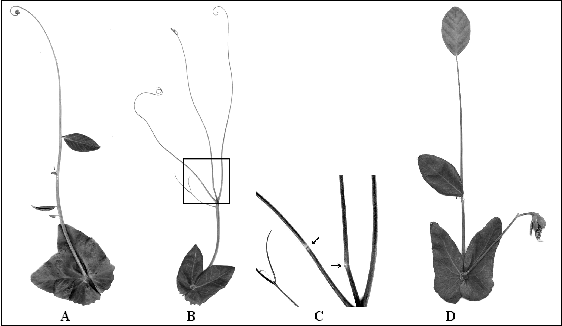
A new allele at Crd disturbs development of the compound leaf
Berdnikov, V.A. and GorelÆ, F.L. and Institute of Cytology and Genetics of Russian Academy of Sciences
Kosterin, O.E. Acad. Lavrentiev ave. 10, Novosibirsk 630090, Russia
Seeds of the SG stock were left to imbibe 0.1% EMS solution and planted. In the M2 population a plant was found with leaflets reduced to a variable extent. Leaflets were sometimes reduced to a hair-like vein a few millimeters long or completely absent, the compound leaf being represented by a pair of stipules and a very thick and long (up to 30 cm) rachis which in its apical zone became a non-branching tendril (Fig. 1A). This plant as well as its descendants often displayed peculiar joints (marked with anthocyanin spots) in the leaf rachis, indicating the nodes from where the missing leaflets would have arisen. The stipules were large, with the margins tending to hang due to weak development of veins. The flowers also exhibited a number of anomalies: the number of sepals often were reduced to 4 or even 3, one or both alae usually were reduced or absent, the elements of the carina often were not fused, and the number of stamens were reduced to 7-8. Due to its resemblance to a whip, we called the new foliage phenotype whip (wh).

Fig. 1. [A] A compound leaf of a pea plant homozygous for the mutation crdwh . [B] A compound leaf of a pea plant with the genotype crdwh / crdwh af/af. The box indicates the area showed in [C] with magnification. [C] Joints (indicated by arrows) on the second order raches of the leaf of a pea plant with the genotype crdwh / crdwh af/af shown in [B]. [D] A compound leaf of a pea plant with the genotype crdwh / crdwh tlw/tlw.
F1 plants resulting from a cross of a mutant plant with the line WL1238 had normal leaves. Among 43 F2 plants eight had phenotype wh, a ratio that does not contradict monogenic nature of inheritance of the mutant phenotype. In an af/af genotype the new mutation decreases the order and intensity of branching (Fig. 1B), with the joints noticeable on the second order raches (Fig. 1C). Especially peculiar was the leaf appearance of the homozygote for the new mutation on the tlw/tlw background: a long rachis (lacking most of its lateral appendages) ending with a large lamina (Fig. 1D).
As a rule, plants possessing the new mutation exhibited reduced fertility, hampering genetic investigations. Nevertheless, after a series of crosses with various lines we obtained a relatively fertile line WHAF homozygous for the mutation and recessive markers a, le and af.
A comparison of wh plants with the mutation crd in the line Wt11300 (obtained from W.K. Sweicicki) revealed a number of similarities. In addition to the parallel in stipular morphology the raches of both mutations often have joints. Also in the flowers of Wt11300 one or both alae are often absent, the carina elements are sometimes free, the stamens as a rule are reduced in number and the fusing of their filaments into a stamen tube is disturbed. F1 plants resulting from a cross WHAF x Wt11300 (a, crd, le, Af), had the whip-phenotype, indicating that the new mutation is a new allele of the locus Crd. We designate the new allele by the symbol crdwh. It should be noted that the allele crdwh disturbs plant development much more strongly than the allele crd, conferring to the pea plant a certain resemblance to Lathyrus aphaca L.
As demonstrated by Swecicki (2) the locus Crd is linked to A and Aatp, located on linkage group II. We obtained F2 progeny from the cross WHAF (crdwh, a, His(2-6)1021, His72) and Yellow Crispa (Crd, A, His(2-6)1221, His73) (the origin of Yellow Crispa is described in ref. 1) and mapped Crd with respect to A and His(2-6) (Table 1). (In this cross the markers gp and cri of linkage group V also segregated but did not show linkage to crd). The data of Table 1 allow the construction of the following map segment:
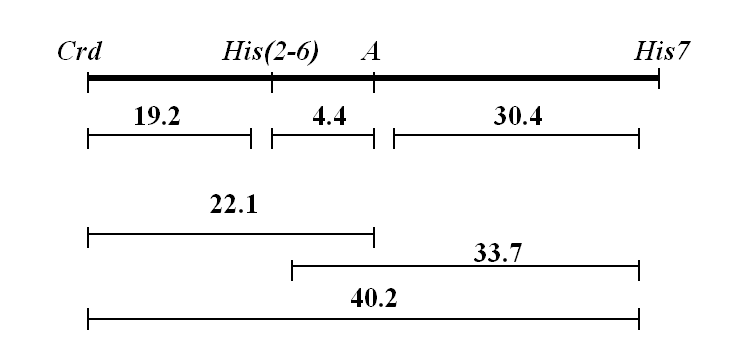
Table 1. Segregation of phenotypes in F2 progeny of the cross WHAF (His(2-6)1021, a, crdwh, His72) x Yellow Crispa (His(2-6)1221, A, Crd, His73). N=268.
|
Gene pair |
Number of plants with designated phenotypes1 |
Joint Chi-sq. |
Recomb fract. (%) |
St. Error |
||||||||
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
|
|
|
|
|
|
|
|
|
||
|
|
|
A/B |
A/h |
A/b |
a/B |
a/h |
a/b |
|
|
|
||
|
a |
crd |
177 |
¢ |
18 |
34 |
¢ |
39 |
61.95*** |
22.11 |
2.94 |
||
|
a |
His(2¢6) |
189 |
¢ |
6 |
6 |
¢ |
67 |
210.87*** |
4.44 |
1.29 |
||
|
a |
His7 |
63 |
105 |
27 |
9 |
31 |
33 |
32.58*** |
30.43 |
3.27 |
||
|
crd |
His(2¢6) |
180 |
¢ |
31 |
15 |
|
42 |
78.80*** |
19.24 |
2.73 |
||
|
crd |
His7 |
59 |
113 |
39 |
13 |
23 |
21 |
8.74 |
40.21 |
3.62 |
||
|
His(2¢6) |
His7 |
60 |
106 |
29 |
12 |
30 |
31 |
23.97*** |
33.70 |
3.40 |
||
1A,a - phenotype
for the first
gene; B,b - for
the second
gene; for codominant alleles in His7
the capital B stands for the allele His73
linked to dominant alleles of other genes; h -
heterozygous; capital
letters stand for dominant alleles,
the haplotype 1221 of the locus His(2-6)
is dominant with respect to its counterpart 1021. Calculations were made using
the maximum likelihood method using the program æCrosÆ.
*,**,*** - probabilities less than 0.01, 0.001 and 0.0001, respectively.
The gene Crd, together with the genes Uni, Af and Tl, clearly are involved in the development of the compound leaf, with Crd apparently being responsible for its branching. Crd also appears to be necessary for normal development of the flower.
Acknowledgement: The work was partly supported by the Russian State Program 'Russian Fund for Fundamental Research', Grant No 99-04-49-970.
1.
Kosterin, O. E., Pukhnacheva, N.V., GorelÆ, F.L., Berdnikov, V.A.
1999. Pisum Genetics 31:
13-20.
2.
Swecicki, W.K. 1989. PNL 21:
73-74.