Double mutant rms2 rms5 expresses a transgressive, profuse branching phenotype
Murfet, I.C. and Symons, G.M. School of Plant Science, University of Tasmania
Box 252-55, Hobart 7001, Australia
Six loci have now been identified in the rms (ramosus) series in pea with up to 11 mutant alleles known at some loci (1, 2, 9, 11, 13, 14, 17). All mutants are recessive and express increased branching at basal and aerial nodes (rms1 to rms5), or at basal nodes only (rms6). The rms mutants are valuable tools for studies aimed at understanding the control of lateral bud outgrowth. These studies have already yielded some valuable insights (4-8, 14-16) and a tentative model now exists for the control of branching in pea by the rms genes (3, 12). This model invokes a role for two unknown and possibly novel messengers.
Determination of the phenotype of rms double mutants can provide additional clues concerning the action and relationships of the rms genes, although the results may not allow unequivocal interpretation. To date, only a small number of rms double-mutant phenotypes have been reported. Two double mutants, rms1 rms2 and rms3 rms6, are known to express a transgressive phenotype (7, 14, 15). In this paper, we report on another double, rms2 rms5, with a transgressive phenotype and several other double combinations where there is no indication of transgression.
Materials and methods
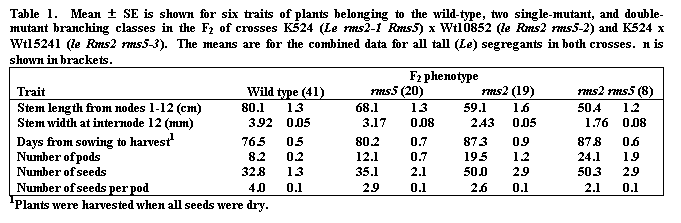
All lines used in the study are held in the Hobart pea collection and details of their origin and nature are given in previous papers (1, 2, 17). Mutant line K524 (rms2-1) was derived from tall (Le) cv. Torsdag and mutant lines Wt10852 (rms5-2) and Wt15241 (rms5-3) were derived from dwarf (le) cv. Paloma. All plants were grown one per 14-cm pot in the glasshouse at Hobart under an 18-h photoperiod obtained by extending the natural day before dawn and after dusk with light from a mixed fluorescent (40 W cool white tubes)/ incandescent (100 W globes) source providing about 25 mmols m-2s-1 at pot top. Nutrient was provided by application of Aquasol (Hortico Ltd, Melbourne) once per week. Details of the traits measured, and branching terminology used, are given in Fig. 1 and legend.
Results
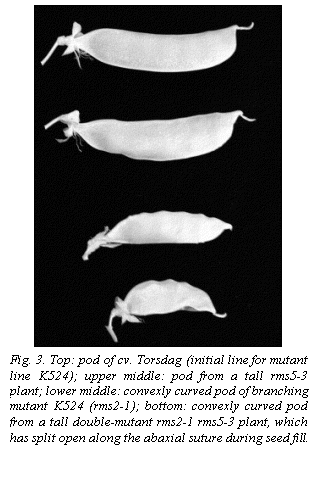
The two crosses K524 (rms2-1) x Wt10852 (rms5-2) and K524 x Wt15241 (rms5-3) gave similar results and combined data have been used in Fig. 2 and Tables 1 and 2. Four phenotypic classes were easily recognisable among tall F2 plants. WT (wild type) plants showed little or no branching and the double-mutant plants showed profuse branching (Fig. 2). The two single mutants displayed an intermediate level of the consistent presence of convexly bent pods like the parental line K524 (Fig. 3). In contrast, the pods on branching (Fig. 2).

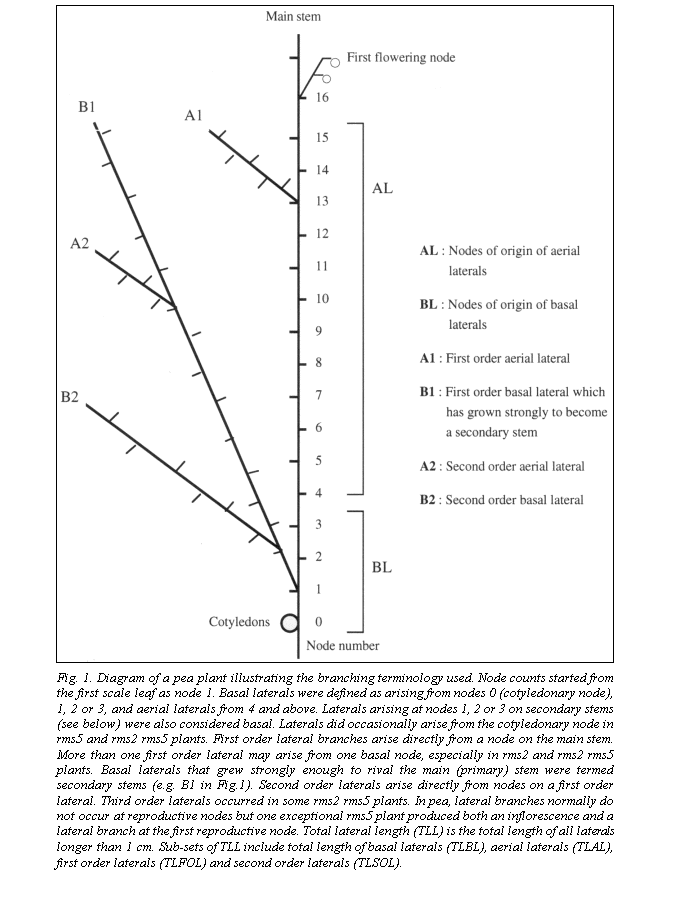
However, rms2 single mutants were distinguishable from rms5 plants by a combination of traits, including shorter, thinner stems (Table 1), a tendency to wilt in sunny conditions and, in particular, rms5 single mutants were generally straight like those of the rms5 parental lines although a few pods on some plants displayed a convexly curved phenotype. The initial line for K524, cv Torsdag, has straight or slightly concave pods (Fig. 3). It is not clear at this time whether the bent pods of K524 are the result of a recessive mutation at a separate locus very close to rms2, or a pleiotropic effect of the rms2-1 mutation itself. To date, we have never obtained a proven recombinant for allele rms2-1 and the bent-pod trait. The bent pods sometimes split open along the abaxial suture as the seeds filled, especially on double-mutant rms2 rms5 plants (Fig. 3).
Four branching phenotypes were also distinguishable among dwarf F2 plants but in this case the separation between the WT and single-mutant phenotypes was less obvious than for tall plants because, like the dwarf WT control, cv. Paloma (initial line for Wt10852 and Wt15241), dwarf WT segregants generally produced some basal and/or aerial laterals.
Segregation at the Rms2 and Rms5 loci was in good accordance with a dihybrid 9:3:3:1 ratio (Table 1, P > 0.2). Seven out of 8 tall putative double-mutant plants were tested by growing F3 (4 plants per progeny) and backcrossing to both single-mutant parents (5-7 seeds of each backcross). All tests gave affirmative results. All backcross plants exhibited a mutant rms phenotype and the F2 plants bred true in F3. The double-mutant F3 plants all branched extensively with the ratio of total lateral length (TLL) to main-stem length in the range of 8-10 (Fig. 2). This ratio is around 3.5-fold greater than for either single-mutant parent thus confirming a strongly transgressive phenotype for the rms2 rms5 double mutant. The two double-mutant F2 plants with TLL to main-stem length ratios of 5.5 and 7.9 (marked with asterisks in Fig. 2) gave rise to F3 progeny with means of 9.0 and 9.2, respectively. Thus the F2 difference appears to reflect the weak early growth of some F2 seedlings (the seed was somewhat old) rather than a difference in genetic potential.
Branching pattern of the F2 plants was influenced by the le mutation for dwarf stature in addition to the rms2 and rms5 mutations. The ability of the le mutation to increase basal branching has been reported before (10) and is clearly seen here in the data for the two single-mutant rms classes (Table 2). The percentage contribution of the basal laterals to total lateral length rose from 47% in tall (Le) rms5 plants to 79% in dwarf (le) rms5 plants (P < 0.01) and the number of basal laterals increased from 1.1 to 2.7 (P < 0.001). A similar effect of le is apparent in the rms2 plants. Among tall plants, the rms2 mutation was slightly more conducive than rms5 to both basal and second order branching (Table 2). All rms plants produced both basal and aerial laterals with the exception of two tall rms5 plants. In general, a gap pattern (see 2) occurred with no laterals arising from nodes immediately above the basal nodes (nodes 0-3). The gap was larger and more clearly defined in rms5 than rms2 plants. The profuse branching of the rms2 rms5 double-mutant plants was reflected in their high level of basal and second order branching (Table 2). Some double-mutant plants produced third order laterals or branched from every main-stem node.
Table 2. Branching pattern for tall (Le) and dwarf (le) rms segregants in the F2 of crosses K524 (Le rms2-1 Rms5) x Wt10852 (le Rms2 rms5-2) and K524 x Wt15241 (le Rms2 rms5-3). The means are for combined data from the two crosses. Basal laterals are defined as arising from the cotyledonary node (node 0) or nodes 1 to 3 on the main and secondary stems. Photoperiod 18 h.
|
Basal laterals as a percentage of total lateral length (%) |
Number of basal laterals longer than 10 cm |
Second order laterals as a percentage of total lateral length (%) |
|||||||||||||
|
Phenotype |
Mean |
SE |
Mean |
SE |
Mean |
SE |
n |
||||||||
|
Tall rms5 |
47 |
6 |
1.1 |
0.2 |
3 |
1 |
20 |
||||||||
|
Dwarf rms5 |
79 |
9 |
2.7 |
0.3 |
9 |
3 |
3 |
||||||||
|
Tall rms2 |
58 |
3 |
2.3 |
0.3 |
18 |
2 |
19 |
||||||||
|
Dwarf rms2 |
76 |
4 |
2.9 |
0.3 |
16 |
4 |
11 |
||||||||
|
Tall rms2 rms5 |
84 |
4 |
9.4 |
1.1 |
23.1 |
2 |
8 |
||||||||
|
Dwarf rms2 rms5 |
83 |
|
7.0 |
|
42 |
|
1 |
||||||||
1Two double-mutant plants produced some small third order laterals that are included in this figure.
Among the four F2 phenotypic branching classes, stem length and width declined in the sequence WT > rms5 > rms2 > rms2 rms5 (Table 1, all differences significant at P < 0.001). For tall double mutants, the total length of the main stem was just under half that of the WT F2 segregants (P < 0.001, data not shown). In addition, both single mutations significantly delayed (P < 0.001) the time when all seeds were dry and ready for harvest, rms5 by 4 days and rms2 by 11 days relative to WT plants (Table 1). The harvest time for the double mutant was similar to that for rms2 plants (Table 1). Mutant plants had more pods and seeds than WT plants but fewer seeds per pod (Table 1) and smaller pods (Fig. 3). These effects were strongest in rms2 and especially rms2 rms5 plants.
The four crosses WL5237 (rms1-1) x K164 (rms4-1), K524 (rms2-1) x K487 (rms3-1), K487 (rms3-1) x K164 (rms4-1), and WL5918 (rms1-3) x WL6042 (rms3-3) gave little or no evidence of transgression. The F2 population of the latter cross contained two plants with branching indices slightly exceeding the parental range, but F3 progeny tests gave no evidence of a genuine transgressive phenotype. In all four F2 populations, segregation was in good accordance with a 9 WT: 7 rms dihybrid ratio. A small F2 was also grown from cross K524 (rms2-1) x K164 (rms4-1). No indication of transgression was obtained but with n = 24 we can only accept this conclusion with about 80% confidence.
Our present results are summarized in Table 3 together with other reported results for rms double-mutant combinations. Seven out of fifteen combinations remain to be tested.
Table 3. Summary of present results and other reported results for crosses segregating at two Rms loci showing the two mutant alleles involved and whether branching in the double mutant falls within the parental range (no transgression) or significantly exceeds the parental range (transgession).
|
|
rms2 |
rms3 |
rms4 |
rms5 |
rms6 |
|
rms1 |
1-1 22 Transg. (7,15) |
1-3 31 No transg. |
1-1 41 No transg. |
|
|
|
rms2 |
|
21 31 No transg. |
21 41 No transg. F2 n = 24 only |
21 52 21 53 Transg. |
|
|
rms3 |
|
|
31 41 No transg. |
|
31 62 Transg. (14) |
|
rms4 |
|
|
|
|
|
|
rms5 |
|
|
|
|
|
Discussion
The four branching mutants rms1, rms2, rms3 and rms4 have now been subjected to fairly extensive study including use of grafting to examine long distance signalling (4-8, 15, 16). Based on these results, Beveridge (3) has suggested that Rms2 may be concerned with the regulation of a novel root-to-shoot signal while Rms1 may be concerned with a second graft-transmissible signal other than cytokinin or auxin moving in a root-to-shoot direction. The action of rms3 and rms4 is not yet clear, but they appear to act largely in the shoot (3, 5). Not enough information is currently available on Rms5 to permit speculation as to its action. Unlike rms1 to rms5, rms6 mutants branch only from the basal nodes, and grafting studies show the primary action of Rms6 may be confined to the shoot (14).
A transgressive phenotype occurred in double mutants rms1 rms2 (7, 15), rms2 rms 5 (Fig. 2, Table 2) and rms3 rms6 (14). The transgression was very marked in the case of rms2 rms5 and occurred with two different alleles, rms5-2 and rms5-3. These results indicate Rms2 probably acts in a different pathway to Rms1 and Rms5, and Rms3 acts in a different pathway to Rms6. However, it should be kept in mind that transgression can occur where two genes act in the same biochemical pathway if both steps are only partially blocked, either as a result of a leaky mutant allele or genetic redundancy for the step.
There was no evidence of transgression in the case of double mutants rms1 rms3, rms1 rms4, rms2 rms3 and rms3 rms4. Likewise for rms2 rms4, but the small numbers here justify only 80% confidence in the result. These five results suggest Rms3 and Rms4 may act in the same pathway. They could also imply Rms1 and Rms2 act in this pathway. However, the transgressive phenotype of the rms1 rms2 double mutant, and evidence from physiological studies (3), indicate that Rms1 and Rms2 act in different ways not only to each other but also to Rms3 and Rms4. This example highlights the fact that while double mutant phenotypes provide useful information on gene interaction, the results can be difficult to interpret with any certainty.
The entire branching control process likely involves a very extensive sequence of events. The branching index used here, ratio of total lateral length to main-stem length, gives only one view of a double mutant. Detailed morphological, physiological and biochemical studies of double mutants may reveal further information on the nature of the interaction. Likewise, molecular analysis of the rms genes could provide crucial information on their nature and likely mode of action.
Acknowledgement: We thank Ian Cummings and Tracey Jackson for technical assistance, Dr. Christine Beveridge and Suzanne Morris for comment on the manuscript, and the Australian Research Council for financial support.
1. Apisitwanich, S., Swiecicki, W.K. and Wolko, B. 1992. Pisum Genetics 24:14-15.
2. Arumingtyas, E.L., Floyd, R.S., Gregory, M.J. and Murfet, I.C. 1992. Pisum Genetics 24:17-31.
3. Beveridge, C.A. 2000. Plant Growth Regulation (in press).
4. Beveridge, C.A., Ross J.J. and Murfet, I.C. 1994. Plant Physiol. 104:953-959.
5. Beveridge, C.A., Ross J.J. and Murfet, I.C. 1996. Plant Physiol. 110:859-865.
6. Beveridge, C.A., Murfet, I.C., Kerhoas, L., Sotta, B., Miginiac, E. and Rameau, C. 1997. Plant J. 11:339-345.
7. Beveridge, C.A., Symons, G.M., Murfet, I.C., Ross, J.J. and Rameau, C. 1997. Plant Physiol. 115:1251-1258.
8. Beveridge, C.A., Symons, G.M. and Turnbull, C.G.N. 2000. Plant Physiol. 123:689-698.
9. Blixt, S. 1976. Agri Hort. Genet. 34:83-87.
10. Murfet, I.C. and Reid, J.B. 1993. In: Casey, R. and Davies, D.R. (eds) Peas: Genetics, Molecular Biology and Biotechnology. CAB International, Wallingford, UK, pp. 165-216.
11. Murfet, I.C. and Rameau, C. 2000. Pisum Genetics 32: 58-59.
12. Napoli, C.A., Beveridge, C.A, and Snowden, K.C. 1999. Curr. Topics Dev. Biol. 44:127-169.
13. Rameau, C., Bodelin, C., Cadier, D., Grandjean, O., Miard, F. and Murfet, I.C. 1997. Pisum Genetics 29:7-12.
14. Rameau, C., Murfet, I.C., Laucou, V., Floyd, R.S., Morris, S. and Beveridge, C.A. 2000. Physiol. Plant. (in preparation).
15. Stafstrom, J.P. 1993. In: Amasino, R.M. (ed.) Cellular Communication in Plants. Plenum Press, New York, pp. 75-86.
16. Stafstrom, J.P. 1995. In: Gartner, B.L. (ed) Physiology and Functional Morphology. Academic Press, San Diego, pp 257-279.
17. Symons, G.M. and Murfet, I.C. 1997. Pisum Genetics 29: 1-6.