Mapping the Rb locus on linkage group III using long PCR followed by endonuclease digestion
| Weeden, N.F. | Department of Plant Sciences |
| Boone, W.E. | Department of Horticultural Sciences Cornell University, Geneva, NY 14456 |
The Rb locus of pea codes for the large subunit of plastid-specific ADP-glucose pyrophosphorylase (1,3). The cDNA sequence coding the large subunit has been published recently (1). We wanted to take advantage of this knowledge of the coding sequence to use the gene as a marker whether or not the rb allele was segregating in the population. The rb allele is a result of a small deletion in the coding sequence, which otherwise is highly conserved. However, numerous introns exist in the gene (2). We suspected that the introns would display greater polymorphism, possibly enough to generate multiple allelic forms within Pisum sativum. We report here the fine mapping of Rb on linkage group III of the most recent pea map (5).
The population consisted of 51 recombinant inbred lines (RILs) from the cross Slow x JI1794 as described in (5). DNA was extracted from each line using the procedure of Torres et al. (4). For PCR amplification, we tested several primers based on the published sequence for the large subunit and determined that the primers PYROLF2 (5Æ-ATG GCT TCT GGT TGT GTG AGC TTG) and PYROLR2 (5Æ-TGC AGT TTC AAG GGA GAG GAT ATA G) produced an approximately 3000 bp fragment that included nearly all the coding sequence and could be conveniently cut with restriction enzymes. The amplification conditions were: 94░ for 60 secs followed by 38 cycles of 94░ for 60 secs, 55░ for 120 secs, 72░ for 240 secs, followed by a 6 min extension at 72░. The reaction buffer, in addition to the standard components contained Taq Extender (Stratagene) and Extender buffer as per manufacturerÆs instructions. The Taq polymerase was purchased from Promega.
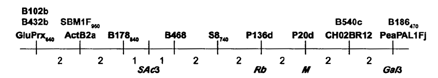
When the amplified fragment was cut with MboI, a difference was observed between the restriction fragment patterns obtained from the two parents. Mapping this polymorphism in the JI1794 x Slow RIL population confirmed the position of Rb between M and the actin gene cluster, SAc3 (Fig. 1). The approximately 2 cM distance between M and Rb must be treated with a degree of skepticism because recombination appears to be somewhat suppressed in this region. Certainly the 4 cM distance between M and Gal3 is significantly shorter than expected from other studies. However, the relative positions of SAc3, Rb, M, and Gal3 are consistent with all previous work.

Fig. 1. Relative locations of markers near M on linkage group III. Numbers below line give approximate map distance in centiMorgans. Markers above line are RAPDs; those below the line are as follows: SAc3 (RFLP), Rb (STS this paper); M (morphological); Ga13 (isozyme).
Although the amplification of such large fragments requires more attention to reaction conditions than standard PCR amplifications, the large fragments appear to contain sufficient DNA sequence polymorphism to permit convenient mapping after testing with a limited number of restriction enzymes. This approach appears to be a general method for mapping sequenced genes containing polymorphic introns.