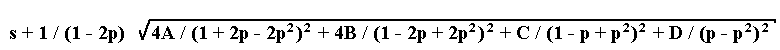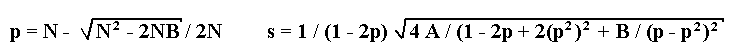
| Kosterin, O.E., Pukhnacheva, N.V., Gorel, F.L. and Berdnikov, V.A. |
Institute of Cytology & Genetics |
We present here equations for estimation
of recombination fraction between a marker and a translocation breakpoint, followed by
pollen fertility versus semisterility, in F2 progenies
with the maximum likelihood method.
If a lethal is linked to a recessive allele a of a marker, then the maximum
likelihood estimation p of the recombination fraction and its standard deviation s will be
determined by the following formulae:
1. A marker is codominant. There exist six phenotypic classes with respect to the marker
and pollen fertility. Let us introduce the following designations: N, the total number of
progeny; A, the sum of three major classes: homozygotes for the marker with fertile pollen
and heterozygote with semisterile pollen; B, the sum of three minor classes: heterozygote
with fertile pollen and homozygotes with semisterile pollen.
Solution of the maximum likelihood equation leads to the following formulae for
estimations of recombination fraction p and its standard deviation s:
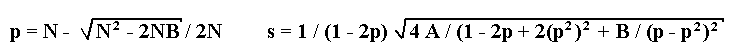
2. The marker is dominant. Let us designate the four phenotypic classes existing as follows: A, dominant fertile; B, recessive fertile; C, dominant semisterile; D, recessive semisterile. The estimation p can be found by numerical solution of the equation
- 2A / (1 + 2p - 2p2) + 2B / (1 - 2p + 2p2) + C / (1 - p + p2) - D / (p - p2) = 0
while the standard deviation is