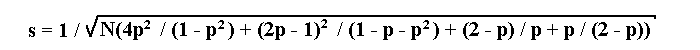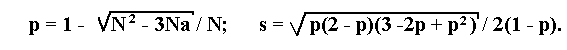
| Kosterin, O.E., Berdnikov, V.A. and Gorel, F.L. | Institute of Cytology and
Genetics |
If a lethal is linked to a recessive allele a of a marker, then the maximum likelihood estimation p of the recombination fraction and its standard deviation s will be determined by the following formulae:
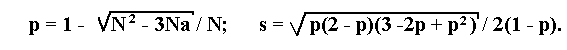
where N is the total number of progeny, a is the number of individuals
with recessive phenotype.
If a lethal is linked to a dominant allele A, then the formulae become:
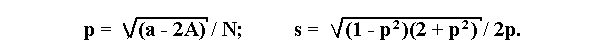
where N is the total number of progeny, A is the number of individuals
with dominant phenotype and a is the number of individuals with recessive phenotype.
If a lethal is linked to an allele B of a co-dominant marker with alleles A
and B then p is to be found by a numerical solution of the following
equation:
2 Np5- (6A +5H +4B)p4+ 2(3A + B)p3+ (5H - 4A + 2B)p2- 2(H + 2B)p + 2B = 0
where N is the total number of progeny, A, B, and H are the numbers of
genotypes A/A, B/B, and A/B, respectively.
The standard deviation s is: