�������� Pisum
Genetics��� Volume 27����� 1995���������� Research Reports���� pages 23-25
Chromosomes 2 and 6 are involved in the Twt-translocation
Temnykh, S.V., Gorel', F.L.,������������������������������������������� Institute
of Cytology and Genetics
and Berdnikov, V.A.��������������������������������������������������������������� Novosibirsk
630090, Russia
Weeden, N.F.������������������������������������������������������������ Department
of Horticultural Sciences
������������������������������������������������������ NYSAES/Cornell
University, Geneva NY 14456, USA
A new translocation closely associated with the
dominant mutation Twt (twisted tendrils) has recently been described (2). Gene Twt was mapped 8 cM from a
(anthocyanin inhibition) in the direction of lf (lower flowering node). Trisomic analysis confirmed that the
interchange chromosome includes the segment
His(2-6) � a � Twt of linkage group IA (2). This linkage
group is believed to belong to chromosome 6 (4, 5), making chromosome 6 a
component of the Twt-translocation. Below we present cytogenetic data that
identify chromosome 2 as the other chromosome
involved in this translocation.
The Twt-line was crossed with known translocation lines from Lamm's
tester set (5). Results of cytological analysis
of the F1 hybrids are given in Table 1.
Table 1. Chromosome
pairing in F1 hybrids between the Twt line and tester interchange
lines.
|
Cross No.
|
Parental Lines
|
Chromosomes of tester line involved in translocation*
|
Chromosome configuration at metaphase I of
meiosis
|
|
No. of bivalents
|
No. of
rings of 4
|
No. of
rings of 6
|
|
1
|
Twt x L84
|
3, 6
|
4
|
0
|
1
|
|
2
|
Twt x L83
|
3, 5
|
3
|
2
|
0
|
|
3
|
Twt x L21
|
3, 4
|
3
|
2
|
0
|
|
4
|
Twt x LI 14
|
1, 2
|
4
|
0
|
1
|
|
5
|
Twt x L108
|
2, 7
|
4
|
0
|
1
|
*Structural types of the tester interchange lines are given according to
Lamm (5) and linkage groups are denoted in
agreement with the version of the pea genetic map published in Vol. 25 of Pisum
Genetics (8).
Results from the first three crosses confirm the involvement of
chromosome 6 in the Twt-translocation (cross
1; Fig. 1 left) and exclude chromosomes 3, 4 and 5 as candidates for the other
member of the interchange (crosses 2 and 3). The presence of one ring of six
and four bivalents in both crosses 4 and 5
(Fig. 1 centre and right) suggests that chromosome 2 is the other chromosome involved in the Twt-translocation.
The structural type of the Twt-translocation
is thus postulated to be T(2,6).
In an attempt to confirm this conclusion we crossed the Twt-line with
lines carrying markers of linkage group VI, the
linkage group currently assigned to chromosome 2. However,
joint segregation analysis (Table 2) gave no significant linkage between Twt
and group VI markers Pl, Prx-3 and Acp-4, and
only a weak linkage between Twt and wlo . The lack of linkage with these standard markers suggests that either the
markers chosen do not cover the entire
chromosome or that the wrong linkage group has been assigned to chromosome 2.
Table 2. Joint segregation data
for Twt and markers of the linkage groups IA, IB and VI and the breakpoint of the translocation Twt.
|
Gene
pair
|
Number of progeny with designated phenotype.1
|
c2
|
Recomb. frac.
� S.E.
|
|
A/B
|
A/h
|
A/b
|
h/B
|
h/h
|
h/b
|
a/B
|
a/h
|
a/b
|
|
a) JI 73 (A, twt, His7F, d, IdhF)
x Twt (a, Tw, His7S,
Dco, IdhS)
|
|
a-Twt
|
2
|
45
|
22
|
-
|
-
|
-
|
21
|
5
|
0
|
63.1****
|
7 � 3
|
|
d-Twt
|
6
|
42
|
6
|
-
|
-
|
-
|
0
|
3
|
16
|
35.8****
|
11 � 4
|
|
His7 - Twt
|
18
|
3
|
0
|
2
|
35
|
6
|
0
|
4
|
11
|
80.1****
|
10 � 3
|
|
Idh - Twt
|
18
|
5
|
1
|
5
|
41
|
6
|
0
|
6
|
15
|
77.9****
|
13 � 3
|
|
a - His7
|
15
|
36
|
5
|
-
|
-
|
-
|
0
|
6
|
14
|
30.5****
|
14 � 4
|
|
a - Idh
|
20
|
42
|
8
|
-
|
-
|
-
|
0
|
9
|
16
|
29.3****
|
18 � 4
|
|
d - His7
|
7
|
30
|
6
|
-
|
-
|
-
|
0
|
6
|
9
|
13.2**
|
21 � 6
|
|
d - Idh
|
9
|
41
|
4
|
-
|
-
|
-
|
0
|
3
|
16
|
41.9****
|
9 � 3
|
|
His7 - Idh
|
9
|
6
|
2
|
4
|
32
|
4
|
0
|
6
|
15
|
47
3****
|
17 � 3
|
|
b) Twt (and, Twt. Acp-4S. Prx-3F)
x Fast (And, twt, Acp-4F,
Prx-3S)
|
|
And - Twt
|
5
|
66
|
27
|
-
|
-
|
-
|
20
|
5
|
1
|
66.0****
|
10 �
3
|
|
Acp-4 - Twt
|
5
|
17
|
3
|
29
|
26
|
16
|
3
|
12
|
13
|
18.0**
|
42 � 4
|
|
Prx-3 - Twt
|
4
|
12
|
9
|
20
|
31
|
20
|
9
|
15
|
4
|
4.4
|
58 � 4
|
|
And - Acp-4
|
30
|
38
|
30
|
-
|
-
|
-
|
2
|
17
|
7
|
7.5*
|
45 � 5
|
|
Prx-3 - Acp-4
|
6
|
18
|
12
|
22
|
29
|
12
|
10
|
16
|
9
|
4.7
|
55 �4
|
|
c) Twt (Twt, Dco, Wlo) x NGB1514 (twt, d, wlo)
|
|
Twt - d
|
296
|
-
|
35
|
-
|
-
|
-
|
38
|
-
|
88
|
163****
|
17 � 4
|
|
Twt - wlo
|
214
|
-
|
35
|
-
|
-
|
-
|
72
|
-
|
26
|
7.6**
|
41 � 4
|
|
d - wlo
|
218
|
-
|
30
|
-
|
-
|
-
|
68
|
-
|
31
|
18.0****
|
36 � 5
|
|
d) Twt (Twt, pl) x K-3953 (twt, Pl)
|
|
Twt - Pl
|
105
|
-
|
31
|
-
|
-
|
-
|
13
|
-
|
5
|
0.2
|
45 � 6
|
1A,a=first gene; B,b=second gene;
h=heterozygous. Where both genes are dominant, the capital letter
stands for the dominant allele. Where the second gene is codominant, a capital
A stands for the dominant allele of the
first gene and capital B for an allele of the second gene which is in coupling
with A. Where both genes are codominant, a capital letter stands for an allele of the first parent. *, **, **** P<0.05,
0.01 and 0.0001, respectively. Data were analysed using the CROS program
developed by S.M. Rozov.
In
contrast to the lack of linkage with group VI markers, Twt displayed
clear linkage with genes on linkage group IB
(d, and, Idh) (Table 2). Although a and d were placed on
the same linkage group on the classical pea map, recent evidence (3, 6)
indicates that the two genes are not on the same chromosome. Paruvangada et al.
(6) found linkage of gene Enod1 with both a on group IA and Fum
on group II and they suggest groups IA and II should be joined. They found no evidence of linkage between Enod2 and
group IB markers. Likewise, in an earlier report, Kosterin (3) failed to show linkage not only between d and
lf
but also between d and the more distally situated group IA loci His7 and blb. Twt maps
in the vicinity of lf and would not be
expected to show linkage with d. However, our data indicate a
recombination value of 10 to
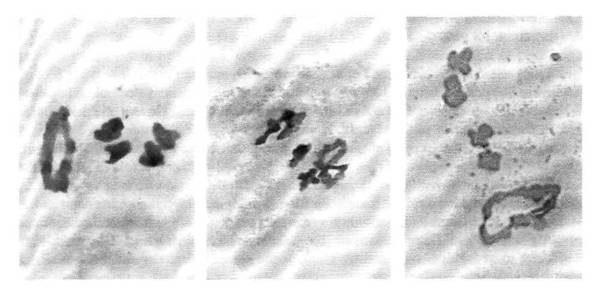
Fig. 1. Chromosomal configurations at the first
metaphase of meiosis in pollen mother cells of F1 hybrids: Left) Twt x L-84; Centre) Twt x L-l14; Right) Twt x L-108.
Acetocarmine staining.
15%
for d and Twt. This apparent contradiction may be explained if 1)
the translocation strongly reduces the recombination in the lower part
of linkage group I or 2) d belongs to the other linkage group involved
in the translocation. The former hypothesis is weakened by our finding that the map distance between His7 and
Twt is comparable to the distance between these loci in the
absence of the translocation (1). Moreover, based on the data from cross JI 73
x Twt, Twt maps equally well between a and His7 and
between His7 and d, indicating pseudo-linkage of the two regions and supporting the latter hypothesis. We also
find a weak linkage observed between d and wlo (Table 2),
suggesting that d may be part of linkage group VI, the linkage group conventionally assigned to
chromosome 2 (7).
However, we do not have compelling evidence for combining linkage group
IB with any of the other linkage groups in
pea. Our results only demonstrate that translocation Twt involves chromosomes
2 and 6, that linkage group IA represents one of the chromosomes (presumably chromosome 6), and that linkage group IB displays
linkage with Twt and what appears to be pseudo-linkage with linkage group IA in crosses involving the Twt
translocation.
1.
Berdnikov, V.A.,
Gorel, F.L., Temnykh, S.V. 1994. Pisum Genet. 26: 9-10.
2.
Gorel', F.L.,
Temnykh, S.V., Lebedeva, I.P. and Berdnikov, V.A. 1992. Pisum
Genet. 24: 48-51.
3.
Kosterin, O.E. 1993. Pisum
Genet. 25: 23-26.
4.
Lamm, R. 1986. PNL 18:
34-36.
5.
Lamm, R. 1987. PNL 19:20-23.
6.
Paruvangada, V.G.,
Weeden, N.F., Cargnoni, T., Yu, J., Gorel', F., Frew, T.,
Timmerman-Vaughan, G.M. and McCallum, J. 1995.
Pisum Genet. 27:12-13.
7. Temnykh, S.V. and Weeden, N.F. 1993. Pisum Genet. 25:1-3.
8.
Weeden, N.F., Swiecicki, W.K.,
Ambrose, M. and Timmerman, G.M. 1993. Pisum
Genet. 25:4 and cover.
