|
Pisum Genetics |
Volume 23 |
1991 |
Research Reports |
pages 14-15 |
Supplemental mapping of the gene Chi-32 on chromosome 3
|
Czerwinska, St., and B. Wolko |
Plant Experiment Station, Wiatrowo 62100 Wagrowiec, Poland |
The chlorotica mutant chi-32 was induced in the line Wt3519 by using fast neutrons for the radiation treatment of dry seeds. The F9 generation analysis of the two point cross Wt11288 (testerline) x WT15325 (mutant) showed linkage between chi-32 and gene M in chromosome 3 (CrO% = 8.5 � 1.7) (1). A recombinant line with genotype chi-32 M was selected from fhis test and included in our Pisum gene bank as the line Wt15327.
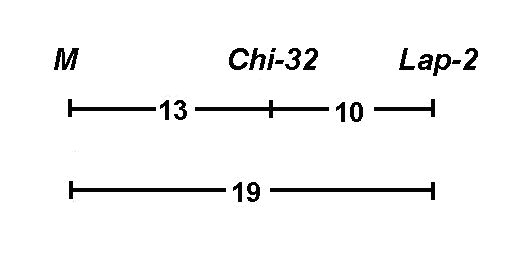
We chose a second marker, the gene Lap-2, for mapping of the chi-32 locus. According to Weeden and Marx (2) the recombination fraction for M - Lap-2 equals 9.0 � 4.9. Line Wt15327 (chi-32 M Lap-2S) was crossed with line Wt8905 (Chi-32 m Lap-2f) and single locus segregation data are shown in Table 1 A. All polymorphisms displayed segregation ratios close to those expected for single gene traits. Joint segregation analysis of chi-32 and markers on chromosome 3 revealed a non random assortment with M (CrO% = 13.7 � 7.1) and with Lap-2 (CrO% = 10.2 � 2.3). The recombination fraction for the pair M - Lap-2 was 18.8 � 7.0 (Table 1 B) This three-point linkage analysis on chromosome 3 indicates the following ranking of the loci:

Our results indicate that the gene Chi-32, which controls a chlorophyll mutation, is placed between M and Lap-2 on chromosome 3. Previous results of Weeden and Marx (2) reveal a smaller distance between M and Lap-2. The cause of this inconsistency is not presently known.
Table
1. Phenotypic distribution in an F2 population derived from cross
Wt8905
(Chi-32 m Lap-2f)
xWt15327 (chi-32 M Lap-2S)
|
A. Monohybrid segregation |
||||
|
Locus |
X |
x |
Total |
Chi-square (3:1) |
|
Chi-32 |
144 |
49 |
193 |
0.01 |
|
M |
136 |
51 |
187 |
0.51 |
|
Lap-2 |
140 |
511 |
191 |
0.29 |
B. Joint segregation of chlorotica chi-32 with M and Lap-2:
|
Gene pair |
XY |
Xy |
xY |
xy |
Total |
Joint |
Rec. fract. |
S.E. |
|
Chi-32 : M |
91 |
51 |
45 |
1 |
187 |
19.77*** |
13.74 |
7.14 |
|
Chi-32 : Lap-2 |
132 |
111 |
8 |
401 |
191 |
105.06*** |
10.23 |
2.33 |
|
M : Lap-2 |
88 |
461 |
49 |
21 |
185 |
17.77*** |
18.88 |
7.03 |
***P< 0.001
1Homozygous slow Lap-2S Lap-2S.
Czerwinska, St. 1988. PNL 20:9-10.
Weeden, N.F. and Marx, G.A. 1984. J. Heredity 75:365-370.