SURFACE SPREAD SYNAPTONEMAL COMPLEXES IN PISUM
SATIVUM
Scheumann, Karin and
Institute of Genetics, University of
Bonn
Gisela Wolf
D-5300 Bonn 1, West
Germany
Surface spreading of meiocytes was
first applied on animal cells (2,3). A similar method was developed for
plant cells (maize) by Gillies (4). Using this method early meiotic
prophase configurations (e.g. pachytene) can be analyzed in a way not
possible by means of conventional methods unless by the use of electron
microscopy and serial sections. Application of the present method destroys
the chromatin leaving for observation only the elements of the
synaptonemal complex. This structure is composed mainly of proteins (see
5) having a special affinity for silver. After staining with
AgN03 the distribution of the silvergrains marks the structure
of interest.
Investigations were carried out on
pollen mother cells of Pisum sativum cv. Dippes Gelbe Viktoria. The
spreading and staining technique for the light microscope as well as for
the electron microscope was done according to Albini and Jones (1) as
modified by Loidl (6) and Loidl and Jones (7).
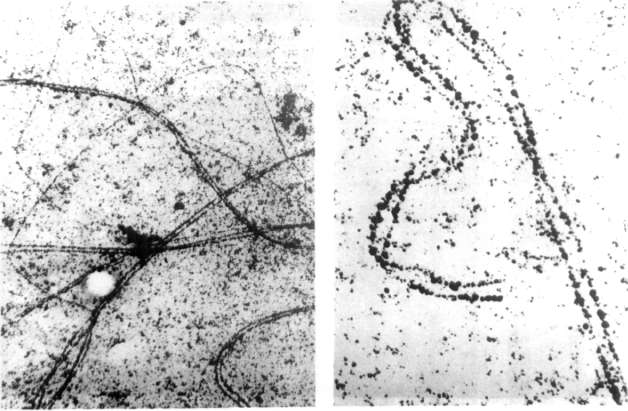
Fig. 1 shows part of a surface
spread zygotene nucleus. Chromatin is not visible, but two parallel
elements of the synaptonemal complex are marked by the accumulation of the
silver grains. The enlargement of the pairing structure (Fig. 2) shows
that it in fact consists of only two elements, namely the lateral
elements, while the central element as well as the transverse elements are
missing. As there is no reason to presume that the synaptonemal complexes
of Pisum sativum differ from the general scheme, we assume that the
central elements and the transverse elements, though present, do not react
with the AgN03 under the applied conditions. Therefore we
conclude that the proteins of the lateral elements and the central
elements have a different amino acid composition. The distance between the
lateral elements measures about 110 nm. This is in full agreement with
data given for other objects.
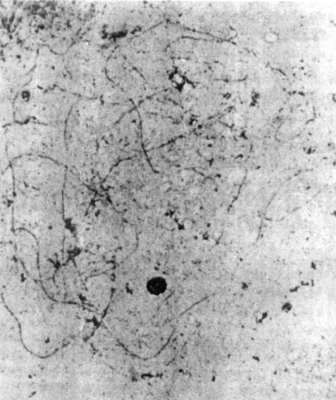
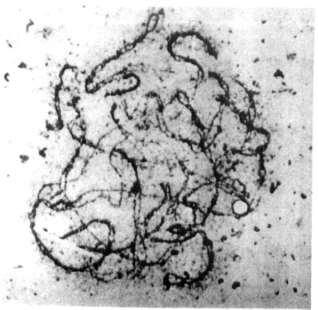
Fig. 3 shows a mid-leptotene stage
in which the axial elements are formed, yet apparently not in
toto but in small pieces, visible as interrupted lineages. Pairing
starts when the axial elements are completed, with alignment beginning
distally (Fig. 4). The more terminal regions are closely related, while
the proximal ones are not. Loops are visible at the very ends in some
bivalents, yet not in all bivalents at the same time. The meaning of these
loops is unknown.
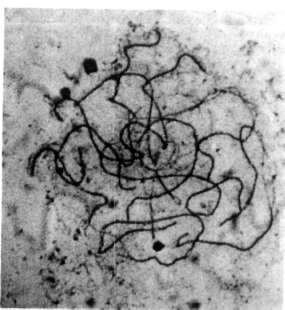
Fig. 5 shows a pachytene stage in
which the bivalents are mostly aligned. Interestingly, no cells were found
in which all bivalents were completely aligned; in each nucleus some
unpaired regions were present.