ELECTROPHORETIC ANALYSIS OF FIELD PEA
CULTIVARS
A. Hussain1, S. T. Ali-Khan2, and W.
Bushuk1
1 - Food Science Dept., University
of Manitoba, Winnipeg, MB, Canada 2 - Agriculture Canada, Research
Station, Morden, MB ROG 1J0, Canada
A procedure was developed to
identify cultivars of field peas (Pisum sativum L.) using
electrophoretic patterns of seed proteins as genotype markers. This paper
presents the details of this procedure.
Eleven cultivars of field peas
registered in Canada ('Bellevue', 'Century', 'Express', 'Fortune',
'Lenca', 'Tara', 'Tipu', 'Titon', 'Trapper', 'Triumph', and
'Victoria') were used for electrophoretic protein analysis. Seeds of each
cultivar were cracked manually using mortar and pestle. Testa-free
cotyledons were ground on a Udy Cyclone mill (Udy Corporation,
Fort Collins, CO), to pass through 1.0 mm mesh screen.
Several different electrophoretic
procedures were tested using extracts of cotyledon proteins; isozymes
were not considered as they do not provide sufficient intervarietal discrimination (2). In the
successful procedure, 0.1 g of seed meal (equivalent to 1/2 - 1. cotyledon) was extracted in
0.45 mL of 5M acetic acid. The
mixture was vortexed for 1-2 min and incubated at 40C tor 2 h. Clear supernatant obtained after cen-trifugation at
8800 x g for 20 min at 23C was
mixed 1:1 with dye solution (3). Ten microliters of the resulting solution
was used for electrophoretic separation. Electrophoresis was carried
out in locally designed acid polyacrylamide gel electrophoresis (PACE)
unit (3). Gels were prepared by
a modification of a previously published procedure (1). Details are
available upon request.
Electrophoresis was carried out
under the following
conditions:
Running buffer
Aluminum lactate, pH 3.1
Running current
15
mA
Running time
6 h
Running temperature 20C After
electrophoresis, the gel was stained overnight in 12% trichloroacetic
acid containing 4% (v/v) Comassie Blue-K solution (3). The stained gel was
rinsed with soapy water, destained 1-4 h,
and photographed.
Reproducibility of the
electrophoregrams was checked by repeating the procedure three times
beginning with the raw seed meal. Results were found to
be highly reproducible.
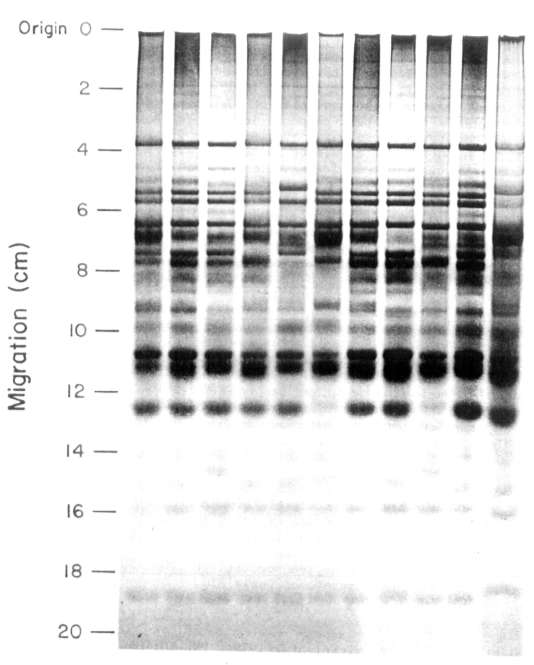
Electrophoretic patterns (or
the 11 cultivars (Fig. 1) were distinctly
different. Electrophoregrams contained 20-23 bands, some of which were genotype
specific while others were present in the patterns of several different
cultivars.
All the cultivars examined
contained one common band. This band can therefore be used as an internal
standard to normalize the data from different gels and to
estimate relative mobilities.
From the results it was obvious
that there were two categories
of bands in the electrophoregrams. First those which are clearly present or absent;
these bands are useful for genotype identification. A second category of
bands were those which were common to several cultivars. These bands may
be valuable in plant breeding if they may be linked to specific agronomic
or quality characteristics.