E. Pagowska, and Institute of Plant Genetics
Polish Academy of Sciences, Poznan, Polanl
N.F. Weeden NYS Agricultural Experiment Station)
Geneva, NY US
Polymorphism in the malate dehydrogenase (L-malate NAD oxido reductase
EC 1.1.1.37; MDH) phenotype has been previously reported in pea (4,5,7) and
attributed to genetic differences among the lines examined. However
genetic tests confirming this assumption have not been published, although
the genetic basis of MDH polymorphism has been established in Zea mays (6),
He1ianthus annuus (1), and Cucurbita pepo (2). In this communication we
present data demonstrating that the variation observed in one region of the
MDH zymogram exhibits Mendelian segregation. Comparison of the MDH
segregation pattern with segregation at other loci indicate that the locus
responsible for the MDH variation is located on chromosome 1.
To obtain the MDH phenotype, leaf extracts were subjected to horlzon-
tal starch gel electrophoresis using a histidine buffer system at pH 6.5 as
described previously (9). The assay solution (25 ml) contained the follow-
ing: 0.1M Tris-HCl pH 8.5, 1 5 mg L-malate, 5 mg NAD, 3 mg MTT and 0.5 mg
PMS. Assays for other isozyme systems have been described (9). The paren-
tal lines used for the genetic analysis were A1078-234, B777-248, A1078-239
(all obtained from Dr. G.A. Marx) and PI 358612 (obtained from the NW
Regional Plant Introduction Station at Geneva).
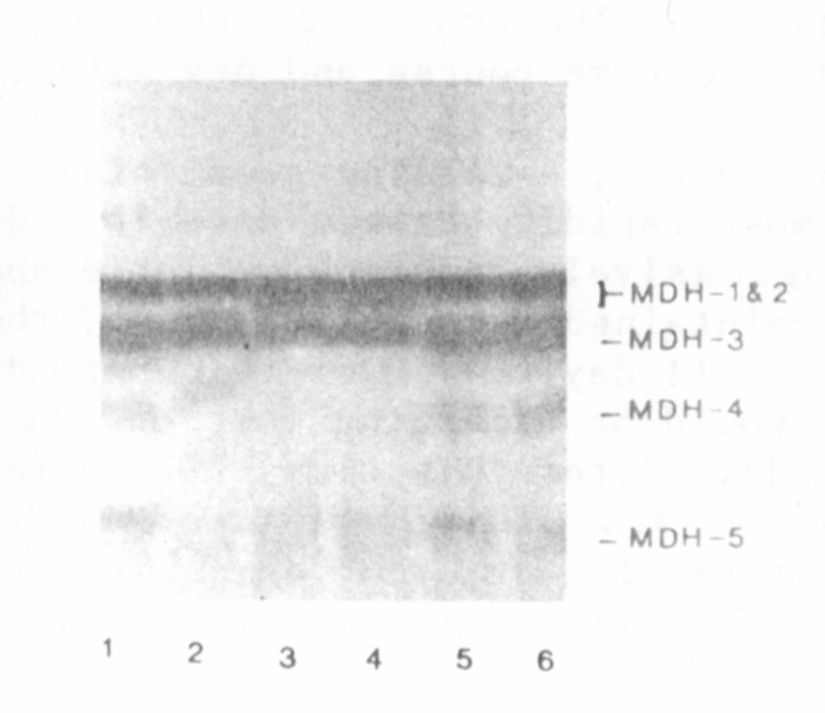
The variation observed in the MDH phenotype is shown in Fig. 1. Line
PI 3586 12 (and B777-248) displayed an MDH band with an R of about 0.25
(MDH-4 in Fig. 1), whereas the other 2 lines lacked this band. Crosses
between lines possessing the MDH-4 isozyme and those lacking such a band
gave Fl plants exhibiting a discernable, although often slightly less
intense, MDH-4 band. In segregating F2 populations we did not attempt to
differentiate between the heterozygous and homozygous MDH-4 "plus"
phenotypes but divided the plants into 2 categories, those exhibiting the
MDH-4 band and those lacking it.
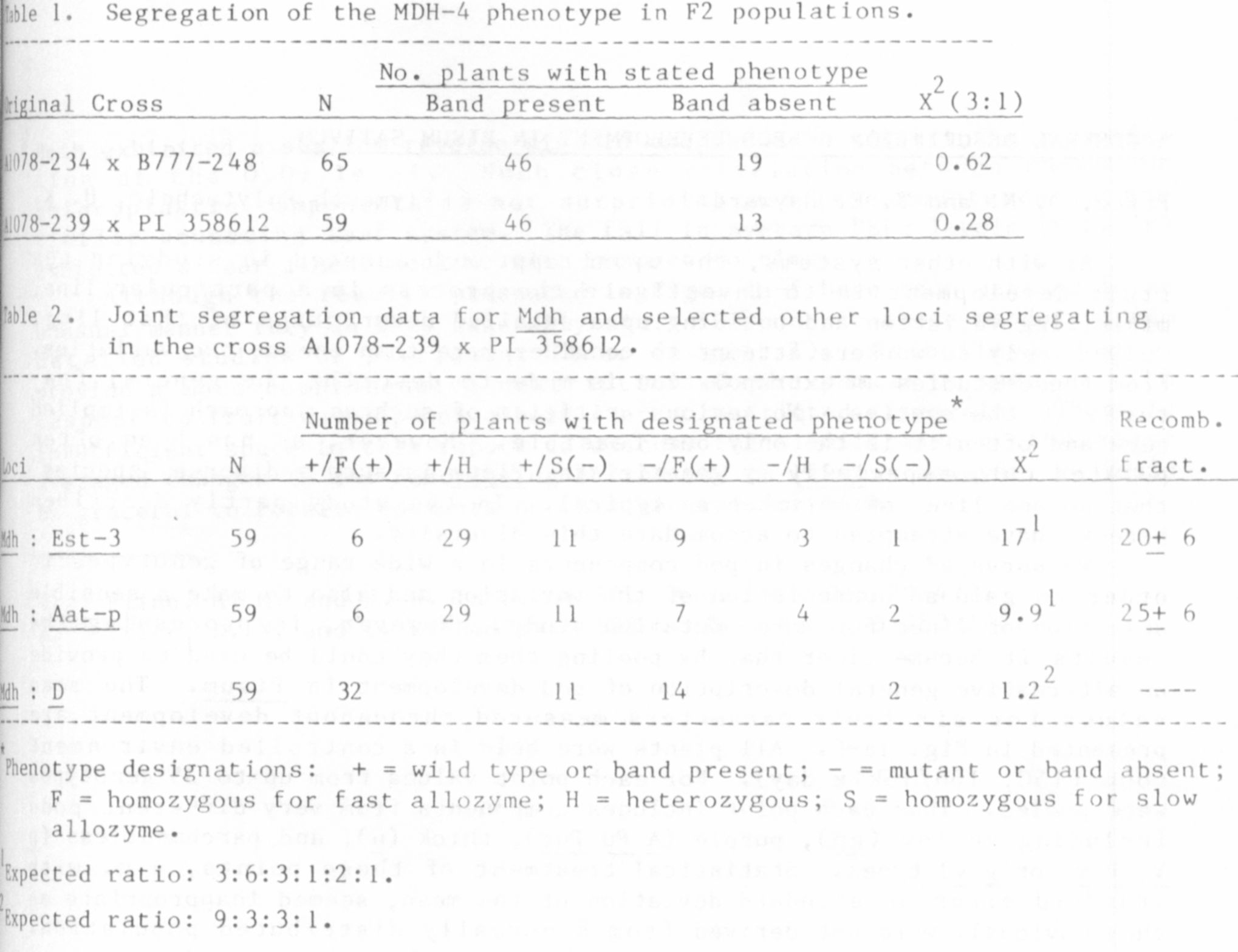
The MDH segregation data from 2 different crosses closelv fits a 3:1
ratio expected for monogenic control (Table !). We suggest that this locus
tentatively be designated Mdh. There are probably other loci coding MDH
isozymes but further characterization of the MDH system is required before
more appropriate locus designations can be made.
Joint segregation analysis revealed non random assortment between Mdh
and 2 isozyme loci on chromosome 1 (Table 2). Both Aat-p and Est-3 have
been mapped near A (3,8) and our data indicate that Mdh also should display
linkage with this morphological marker. Mdh segregated independently of D
Idh, and I, the other chromosome 1 markers segregating in one or both of
the crosses listed in Table 1 (data not shown).