68 RESEARCH REPORTS
ONL Volume 12 1980
INHERITANCE OF PROTEIN CONTENT IN PEA
II. HERITABILITY OF PROTEIN CONTENT IN RANGER X STRAL AND PALOMA X STRAL CROSSES
Swiecicki, W.K., Z. Kaczmarek, Plant Experiment Station, Wiatrowo, Poland
and M. Surma Institute of Plant Genetics, Poznan, Poland
On the basis of breeding data obtained in the past, three varieties were
chosen for studies, viz. 'Ranger1, 'Stral' and 'Paloma'. The breeding data
showed some strains with a higher protein content (WTD 4011 and WTD 4015),
originating from crosses where one of the parents was the variety Ranger or
Stral. Paloma lias a low protein content, but is among the highest yielding
varieties in Poland. Crosses were made between varieties Ranger x StrSl and
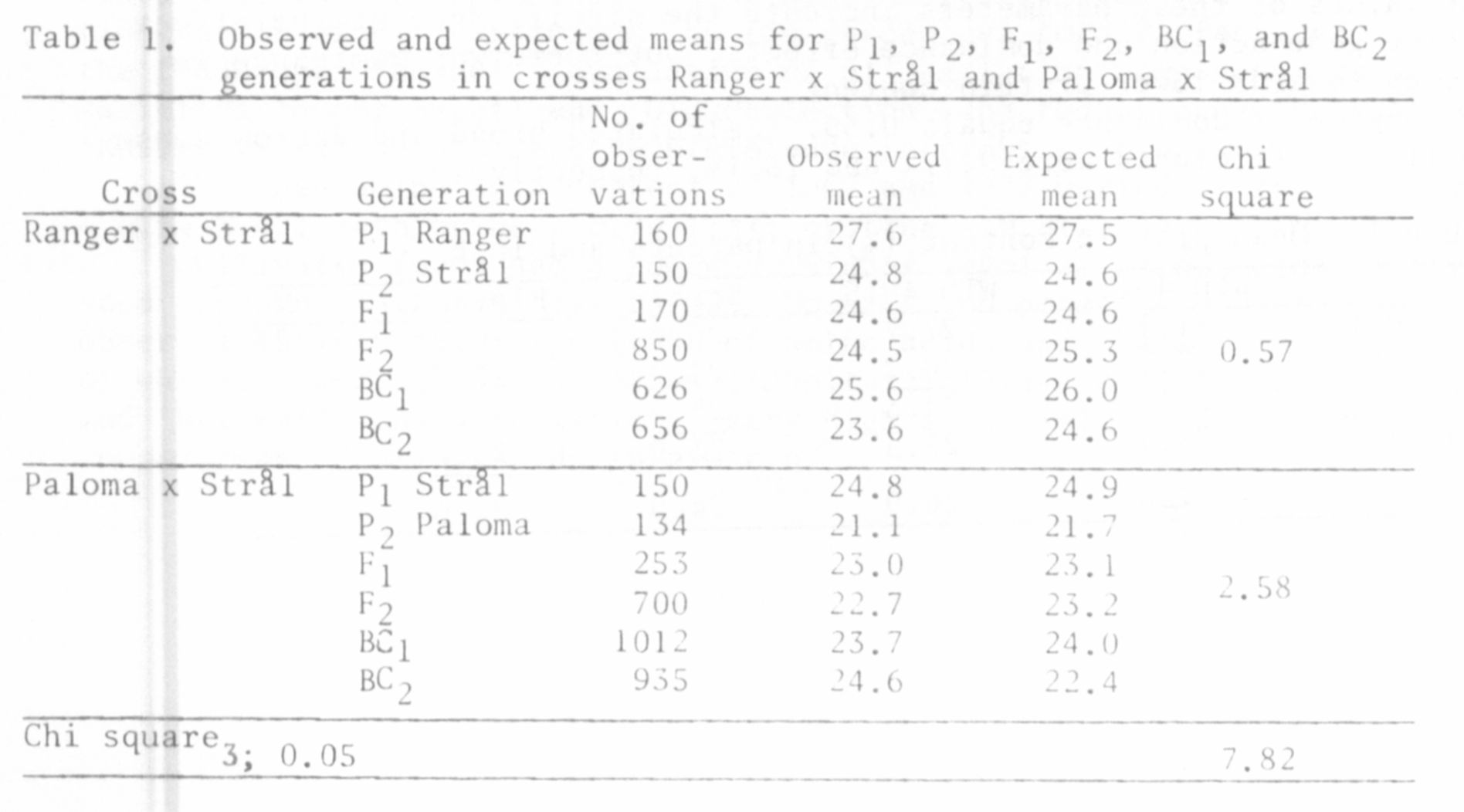
Paloma x Stral. The F1, F2, BC1, BC2, and parents were tested in each cross.
Protein content was analyzed in single seeds by the Kjehldal method.
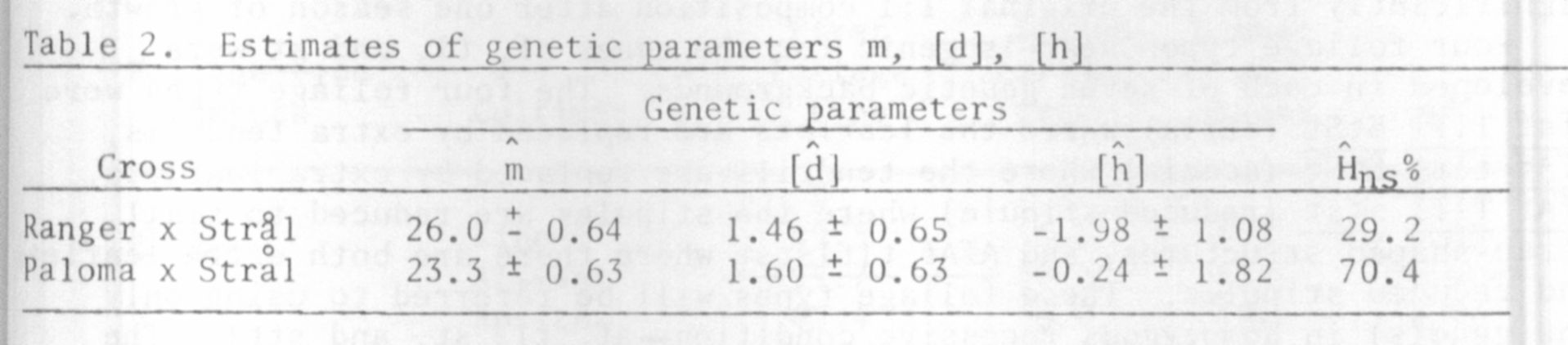
To assess the degree of genetic control over the protein content in the
seeds, heritability coefficients were estimated in a narrow and a broad sense.
The heritability coefficient estimate in the broad sense was calculated by
a commonly used formula, i.e. by estimating the variance of the F2 and that
of the parents. The value of this coefficient for the cross Ranger x Stral
was 79.8% and that for Paloma x Stral 78.0%.
From the point of view of breeding it is far more interesting to know
the heritability coefficient estimate in the narrow sense, expressed by the
additive to phenotypic variance ratio (Mather and Jinks, 1971). It is possible
to estimate this coefficient when gene action in a given cross is additive-
dominance (Kaczmarek, Surma, and Swiecicki, in press). The adequacy of the
additive-dominance model of gene action was tested using the significance
of differences between the observed and expected means for the analyzed genera-
tions (Chi square statistics). The results are presented in Table 1.